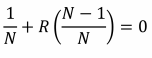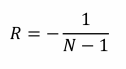Alex Tep
A more technical post. It will be helpful to be familiar with the idea of the ‘selfish gene’ in evolutionary biology before reading this.
Although our treatment involves E. coli – a species of bacteria that naturally occurs in the gut – creating a stable population of engineered bacteria in the gut will be tricky. Even if our E. coli are able to establish themselves in the gut, they must compete with native gut E. coli for the exact same resources. This is a big problem – our engineered bacteria must allocate some resources that could be used for reproduction to producing the copper chelator. Meanwhile, since native E. coli can commit all their resources to reproduction, they will necessarily be better at reproduction than our E. coli, and will gradually become better represented in the gut than the treatment bacteria until our bacteria are totally expunged.
To prevent this from happening, we are arming our bacteria with bacteriocins – proteins that kill other bacteria. This will allow our E. coli to kill its native rivals, preventing them from being outcompeted. But to release this toxin, E. coli cells must burst open suicidally. How could natural selection favour a gene for a trait that harms its bearer? Well, if the recipient is related to the bearer, it will have a greater than average chance of sharing the same gene as the bearer; the closer the relation, the larger this probability. So if an individual sacrifices itself to save the lives of several close relatives (and the copies of the altruistic gene carried by them), although one copy of the gene is lost, several are saved. Over time, this gene, and the trait it encodes, can become stable in the population. But explaining the existence of traits that harm both actor and recipient, like bacteriocins – well, that is more difficult.
Darwin made pioneering successes in explaining adaptation, but he struggled with behaviours that harm the individual performing them. Regarding altruism, traits that reduce an individual’s own reproductive success – or fitness – to raise another’s, he wrote:
‘He who was ready to sacrifice his life, as many a savage has been, rather than betray his comrades, would often leave no offspring to inherit his noble nature.’ (Darwin, 1871)
His solution to this paradox was to look at the effect of altruism on the group of which the individual is a part.
‘A tribe including many members who… were always ready to give aid to each other and sacrifice themselves for the common good, would be most victorious over most other tribes; and this would be natural selection.’ (Darwin, 1871)
But this group-selectionist view of altruism fell out of favour in the twentieth century: altruistic groups were too vulnerable to subversion by selfish cheaters. A free-riding individual joining the tribe that received this aid from other members but refused to help them in return would increase in fitness. And their offspring, who inherit their selfish nature, would increase in frequency at the expense of their comrades’, until there are no altruistic individuals left.
In a series of influential papers, W D Hamilton demonstrated that there is no need to invoke group selection to explain altruistic traits (Hamilton, 1964). When natural selection acts on genes rather than groups, it is possible for a gene for an altruistic trait to increase in frequency if the altruism is directed at individuals that share this gene. More formally, this can be expressed as Hamilton’s rule, which states that selection can act on and favour genes for altruistic traits when:
rB > C
(1)
or when the benefit to the recipient of the action (B), adjusted by its relatedness to the actor (r) – loosely, the likelihood that the recipient shares such a gene, outweighs the cost to the actor (C). This emphasised the value of considering the fitness of relatives as well as that of the focal individual– inclusive fitness – when thinking about social traits.
But as well as explaining altruism, Hamilton’s rule carries a darker suggestion: that it is possible to satisfy the inequality rB > C or, more generally rB – C > 0, when C is positive and B is negative. Where altruism is helping another at personal cost, spiteful traits harm both actor and recipient (Hamilton, 1970). Explaining how natural selection could possibly favour such mutually destructive qualities has been more problematic.
Firstly, the keen-eyed will notice that in order for Hamilton’s rule to be satisfied in cases of spite, if B is negative and C is positive, r must also be negative:
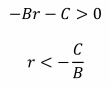 (2)
(2)
But how is it possible to be negatively related to someone? True, if we think of relatedness as the chance of sharing a gene, r cannot be negative because it is a probability. But Hamilton’s rule is much more useful to us if we think of relatedness as a regression coefficient (which can take negative values) of the recipient’s genes on the actor’s (Hamilton, 1963). In his geometric view of relatedness, Grafen (Grafen, 1985) emphasises the value of considering the genetical composition of the population: if the recipient of a social trait carries the actor’s genes at a frequency p, that is greater than the genes’ frequency in the population 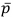 , increased reproductive success of the recipient means an increased frequency of the actor’s genes in the population, and thus a gain in the actor’s inclusive fitness (RB > 0 (Eqn. 1)). If the recipient carries the actor’s genes at a frequency lower than their frequency in the population, an increase in the recipient’s reproductive success means a reduction in the frequency of the actor’s genes in the population, and thus a loss in inclusive fitness for the actor (RB < 0 (Eqn. 1)). And if p =
, increased reproductive success of the recipient means an increased frequency of the actor’s genes in the population, and thus a gain in the actor’s inclusive fitness (RB > 0 (Eqn. 1)). If the recipient carries the actor’s genes at a frequency lower than their frequency in the population, an increase in the recipient’s reproductive success means a reduction in the frequency of the actor’s genes in the population, and thus a loss in inclusive fitness for the actor (RB < 0 (Eqn. 1)). And if p = 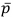 , relatedness is zero. This is important because changing the value of
, relatedness is zero. This is important because changing the value of 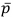 alone can change the outcome of inclusive fitness for the actor. So negative relatedness simply means that the recipient’s relatedness to the actor is less than its expected relatedness to a random member of the population.
alone can change the outcome of inclusive fitness for the actor. So negative relatedness simply means that the recipient’s relatedness to the actor is less than its expected relatedness to a random member of the population.
Second, though negative relatedness is possible, we can normally only expect relatedness between an actor and its social partner to be only slightly negative – so slight as to be considered a trivial force in Hamilton’s rule. An individual living in a population of size N is related to itself, a fraction 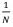 of the population, by amount 1, and to the rest of the population,
of the population, by amount 1, and to the rest of the population,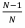 , by amount R. As above, relatedness to the population as a whole must be zero, so:
, by amount R. As above, relatedness to the population as a whole must be zero, so:
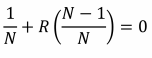
Rearranging gives us the average relatedness between the actor and its social partners:
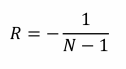
(3)
If the population is of any substantial size, N will be large enough to make R only weakly negative. For this reason, Hamilton himself did not place much importance on spite because of its restrictive requirements – populations would have to be tiny for R to be negative enough to have any effect – in his words, spite is merely the ‘final infection that kills falling twigs off the evolutionary tree’ rather than an significant biological phenomenon (Hamilton, 1996).
Indeed examples of spite have been rare, and many of those that have been found actually turned out to be examples of selfishness – actions that harm the recipient to increase the actor’s inclusive fitness. For example, that herring gulls commit siblicide on chicks at neighbouring nests has been cited as an example of spite (Pierotti, 1980), but since this action reduces competition for themselves and their offspring in the future, this is really selfishness (Foster, Wenseleers and Ratnieks, 2001). As West and Gardner put it: ‘[w]hat matters for natural selection are fitness consequences over the entire lifetime and not some arbitrary period’ (West and Gardner, 2010).
But while populations may be large, social interactions do not always occur on a global scale. When looking at a much smaller, more local social arena – for example in the presence of local competition, R can be sufficiently negative to allow for spiteful traits.
Further, the magnitude of R can be boosted if the actor is able to discriminate between closer and more distant relatives. How could this ability arise? Suppose there arose a gene that conferred some conspicuous phenotype, say, for a green beard, and the ability to recognise such a trait and treat bearers of this trait preferentially. This greenbeard gene would increase in frequency in the population by directing altruism towards carrier-relatives and spite towards non-relatives.
E O Wilson has disputed the necessity of negative relatedness altogether for the evolution of spite. He suggested that spite towards non-negatively related individuals could be favoured if the action helped a relative to a more-than-compensatory degree (Wilson, 1976). A modification to Hamilton’s rule can incorporate a third-party relative:
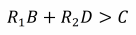
(4)
where R1 is the relatedness to the recipient, R2 is the relatedness to the third-party relative and D represents any benefits enjoyed by this relative. Using this model, Wilsonian spite can occur when D > 0 and R2 > 0, even if R1 > 0. Contrast this to Hamiltonian spite where R1 < 0 and R1B > 0 and D is not necessarily > 0 (Gardner and West, 2004).
Then isn’t spite just altruism to a third-party? Why is it useful to distinguish this from altruism? There are obvious differences between spiteful and altruistic traits from a behavioural perspective, but there are some biologically interesting differences too. We expect a positive linear relationship between fitness and relatedness for altruistic traits – the more closely related the actor is to the recipient, the more likely that the recipient will contain the gene for the trait in question, and the more favourable altruism towards the recipient is.
But with spiteful traits mathematical models predict a domed relationship between fitness and relatedness: when the fraction of spiteful individuals is low, 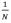 is low and that of non-relatives,
is low and that of non-relatives, 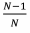 (Eqn. 3) is high, making R only weakly negative (Eqn. 3) and insufficient to outweigh the cost of the spiteful trait. When the focal group dominates the population, there are too few non-relatives for the harm to the recipient to outweigh the cost to the individual. Kin discrimination is often key in providing negative relatedness for Hamiltonian spite – altruism can evolve without kin discrimination if, for example, dispersal is limited so that relatives stay together.Also, we can expect spite, rather than altruism to be favoured when there is local competition since this is a source of negative relatedness.
(Eqn. 3) is high, making R only weakly negative (Eqn. 3) and insufficient to outweigh the cost of the spiteful trait. When the focal group dominates the population, there are too few non-relatives for the harm to the recipient to outweigh the cost to the individual. Kin discrimination is often key in providing negative relatedness for Hamiltonian spite – altruism can evolve without kin discrimination if, for example, dispersal is limited so that relatives stay together.Also, we can expect spite, rather than altruism to be favoured when there is local competition since this is a source of negative relatedness.
Observations on bacteriocin-producing bacteria are consistent with the theory. Bacteriocins are antimicrobial compounds secreted by a wide variety of bacteria (Riley and Wertz, 2002). They are costly to produce (cell lysis is required for bacteriocin release in Escherichia coli, but there is a substantial metabolic cost in all producers) and they kill competing susceptible cells through enzyme inhibition, membrane pore formation, and their nuclease activity, among other mechanisms (Riley and Wertz, 2002); so bacteriocin release can be considered to be a spiteful trait. One might argue that, in non-autolysing cells, the trait is selfish rather than spiteful since the individual gains through loss of competition by killing neighbouring susceptible, but bacteriocins are highly diffusible and an actor is unlikely to benefit from the death of a distant competitor (Inglis et al., 2009). Crucially, bacteriocin-production can be considered a greenbeard phenotype since bacteriocins specifically target non-related individuals while leaving relatives untouched because immunity factors are often linked to the toxin gene (Riley and Wertz, 2002).
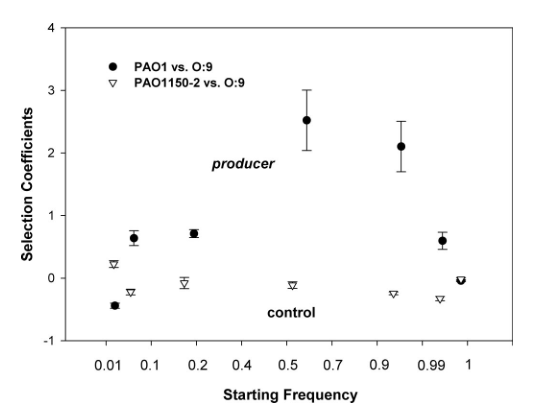
Inglis et al. used Pseudomonas aerugnosa and a caterpillar model to demonstrate the importance of relatedness in allowing for spiteful traits (Inglis et al., 2009). In vitro studies competed a bacteriocin-producing strain with a susceptible competitor and the initial frequency of the producing strain was manipulated and its fitness measured as growth rate relative to the susceptible strain. At low and high starting frequencies, producer fitness was low, and fitness was greatest at intermediate starting frequencies (Fig. 1a). In vivo studies in the caterpillar gave comparable results. Infecting populations were manipulated to contain a high (99%), intermediate (50%), or low (1%) frequency of the producing strain relative to the susceptible. As expected, caterpillars inoculated with the intermediate population took significantly longer to die than those treated with the high and low populations (Fig. 1b).
1a.
1b.
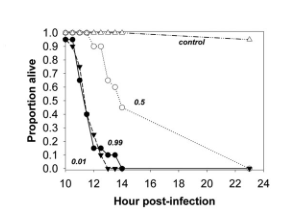
FIG. 1 (Inglis et al., 2009)
At high producer frequencies, there are only a few competitors to kill, and thus only a few resources to gain by releasing bacteriocin. The increase in inclusive fitness is insufficient to counterbalance the cost of bacteriocin release. At low producer frequencies R is only weakly negative and the impact on susceptible bacteria is negligible and insufficient to outweigh the costs of bacteriocin production.
Is this an example of Hamiltonian or Wilsonian spite? That there are elements of both here suggests that they are just different ways of looking at the same thing: Wilson just used a different reference population to calculate relatedness so that it was not negative. Indeed, Wilson himself did not draw a distinction between the two, his figure in Sociobiology (Fig. 2) illustrates Hamilton’s view of spite, and more careful inspections of the mathematics of both has demonstrated their synonymy.
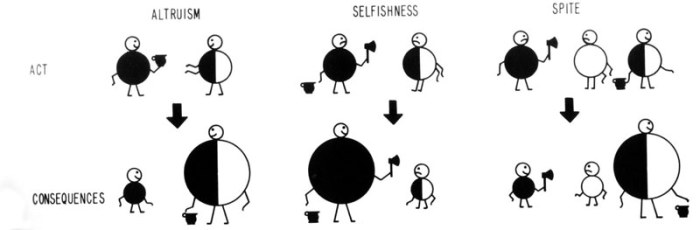
FIG. 2 (Wilson, 1976)
Behaviours that harm the individual performing them have long remained a problem for evolutionary theory. But even with Hamilton’s explanation of altruistic behaviour, it has often been assumed that spite cannot be favoured for its restrictive requirements. This was to neglect the importance of local competition and a growing cache of examples exists where kin discrimination, negative relatedness and third-party altruism interact to allow for spiteful behaviour.
REFERENCES
Bourke, A.F.G. (2011) Principles of Social Evolution, 1st edition, Oxford: Oxford University Press.
Darwin, C. (1859) On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life, 1st edition, London: John Murray.
Darwin, C. (1871) The descent of man, and selection in relation to sex, London: John Murray.
Davies, N.B., Krebs, J.R. and West, S.A. (2012) An Introduction to Behavioural Ecology, 4th edition, Oxford: Wiley-Blackwell.
Fisher, R.A. (1930) The Genetical Theory of Natural Selection, Oxford: Clarendon.
Foster, K., Wenseleers, T. and Ratnieks, F. (2001) ‘Spite: Hamilton’s Unproven Theory’, Ann. Zool. Fennici, vol. 38, pp. 229-238.
Gardner, A. and West, S.A. (2004) ‘Spite and the scale of competition’, Journal of Evolutionary Biology, vol. 17, pp. 1195-1203.
Grafen, A. (1985) ‘A geometric view of relatedness’, in Dawkins, R. and Ridley, M. Oxford surveys in evolutionary biology, Oxford: Oxford University Press.
Hamilton, W.D. (1963) ‘The evolution of altruistic behaviour’, American Naturalist, vol. 97, pp. 354-356.
Hamilton, W.D. (1964) ‘The Genetical Evolution of Social Behaviour. I’, Journal of Theoretical Biology, vol. 7, February, pp. 1-16.
Hamilton, W.D. (1970) ‘Selfish and spiteful behaviour in an evolutionary model’, Nature, vol. 228, pp. 1218-1220.
Hamilton, W.D. (1996) Narrow Roads of Geneland Volume 1: Evolution of Social Behaviour, Oxford: Freeman.
Inglis, R.F., Gardner, A., Cornelis, P. and Buckling, A. (2009) ‘Spite and virulence in the bacterium Pseudomonas aeruginosa’, PNAS, vol. 106, no. 14, April, pp. 5703-5707.
Pierotti, R. (1980) ‘Spite and altruism in gulls’, American Naturalist, vol. 115, pp. 290-300.
Riley, M.A. and Wertz, J.E. (2002) ‘Bacteriocins: evolution, ecology, and application.’, Annu. Rev. Microbiol, vol. 56, pp. 117-137.
West, S.A. and Gardner, A. (2010) ‘Altruism, Spite, and Greenbeards’, Science, vol. 327, March, pp. 1341-1344.
Wilson, E.O. (1976) Sociobiology: The New Synthesis, 1st edition, Cambridge: Belknap Press of Harvard University Press.
 sented their idea in the Bazaar at the London International Youth Science Forum (LIYSF), which took place from 27th July to 10th August. LIYSF celebrates its 58 years of history, and has been granted UNESCO patronage this year. The forum involves lectures from pioneering researchers, visits to research centres in the UK, and a rich engagement and exchange of ideas amongst participants. 475 representatives from 75 countries participated, and over a hundred science projects were presented.
sented their idea in the Bazaar at the London International Youth Science Forum (LIYSF), which took place from 27th July to 10th August. LIYSF celebrates its 58 years of history, and has been granted UNESCO patronage this year. The forum involves lectures from pioneering researchers, visits to research centres in the UK, and a rich engagement and exchange of ideas amongst participants. 475 representatives from 75 countries participated, and over a hundred science projects were presented. finding alternative treatments for Wilson’s disease and raising public awareness towards rare disease. Most of the participants were unaware of Wilson’s disease, and they showed interest towards the issue. One student from Poland, however, was diagnosed with Wilson’s disease, which was later proven to have been false-positive. He showed genuine interest towards this issue, being aware of current treatments, and was keen to see improvements on both treatment and diagnosis.
finding alternative treatments for Wilson’s disease and raising public awareness towards rare disease. Most of the participants were unaware of Wilson’s disease, and they showed interest towards the issue. One student from Poland, however, was diagnosed with Wilson’s disease, which was later proven to have been false-positive. He showed genuine interest towards this issue, being aware of current treatments, and was keen to see improvements on both treatment and diagnosis.



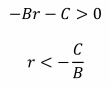 (2)
(2)